Skills








Experience
Graduate Researcher, Lehigh University
Bethlehem, PA
My doctoral research focuses on developing computational methods to study protein–protein interactions at the level of individual residues and intermolecular bonds. Rather than treating protein complexes as abstract inputs, my work models interaction interfaces as structured networks, enabling mutation-level analysis of binding affinity and interaction mechanisms. This research combines model development, dataset construction, and software tooling to support interpretable and reproducible molecular science.
- Designed graph-based representations of protein interfaces that explicitly model hydrogen bonds, ionic interactions, salt bridges, and residue contacts.
- Developed graph neural network architectures tailored to these representations to analyze mutation-driven changes in binding affinity.
- Constructed annotated datasets linking experimental mutation data to interaction mechanisms described in the literature.
- Built visualization tools and web servers that map between model outputs and three-dimensional protein structures.
- Resulted in six peer-reviewed publications across leading bioinformatics venues, including a Best Paper Finalist at IEEE CSBW.

AI Graduate Researcher, Los Alamos National Laboratory
Los Alamos, NM
At Los Alamos National Laboratory, I worked on automating the analysis of large ensemble scientific simulations that generate terabytes to petabytes of data. The goal of this project was to make complex simulation outputs easier to explore while preserving full transparency and reproducibility. This work resulted in InferA, a provenance-aware system for structured data analysis and visualization used in national lab research workflows.
- Designed and implemented InferA, a system for automating analysis and visualization of large-scale cosmology simulation ensembles.
- Built modular multi-agent workflows to coordinate data filtering, statistical analysis, and visualization tasks.
- Integrated LangChain and LangGraph to support schema-aware querying, tool calling, and intelligent task delegation.
- Implemented full provenance tracking so every result can be traced back to source data and parameters.
- Evaluated scalability, modularity, and interpretability to inform future scientific AI infrastructure at LANL.
- Published this work at SC’25 (AI4Science).

Bioinformatics Researcher (Co-op), Moderna Therapeutics
Cambridge, MA
At Moderna, I worked on computational approaches for designing human-like mRNA sequences with improved stability and controllable properties. The project focused on bridging structural information and sequence generation to overcome limitations in existing mRNA design methods. This work contributed models and tooling that were integrated into active research pipelines.
- Developed transformer-based generative models for human-like mRNA sequence design using PyTorch.
- Built a hybrid architecture that integrates graph neural network features into sequence generation.
- Enabled fine-grained modulation of normally inflexible mRNA properties at single-codon resolution.
- Integrated models and features into production research pipelines used by downstream teams.
- Presented technical results to VP- and director-level research leadership.

Adjunct Professor in Mathematics, NYC College of Technology
Brooklyn, NY
In this role, I taught undergraduate mathematics courses while also contributing to instructional infrastructure used across the department. My teaching emphasized conceptual understanding and active engagement, alongside scalable tools to support consistent assessment.
- Taught Pre-Calculus, Calculus I, and Calculus II to undergraduate students.
- Co-developed a web-based interactive homework and grading platform adopted college-wide.
- Used a mix of lecture-based and activity-driven instruction to improve engagement with challenging topics.

Publications
InferA: A Smart Assistant for Cosmological Ensemble Data
Workshops of the International Conference for High Performance Computing, Networking, Storage and Analysis (SC Workshops ’25), St. Louis, MO, USA
This paper presents InferA, a system designed to make large ensemble cosmology simulations easier to analyze by automating common analysis and visualization workflows while preserving full provenance. The goal is to allow scientists to explore simulation outputs at scale without losing transparency or reproducibility.
InferA interprets natural language queries to coordinate modular agents that perform schema-aware data querying, statistical analysis, and visualization on terabyte- to petabyte-scale datasets. The system tracks every intermediate result and transformation, enabling results to be audited, reproduced, and extended across large scientific workflows.
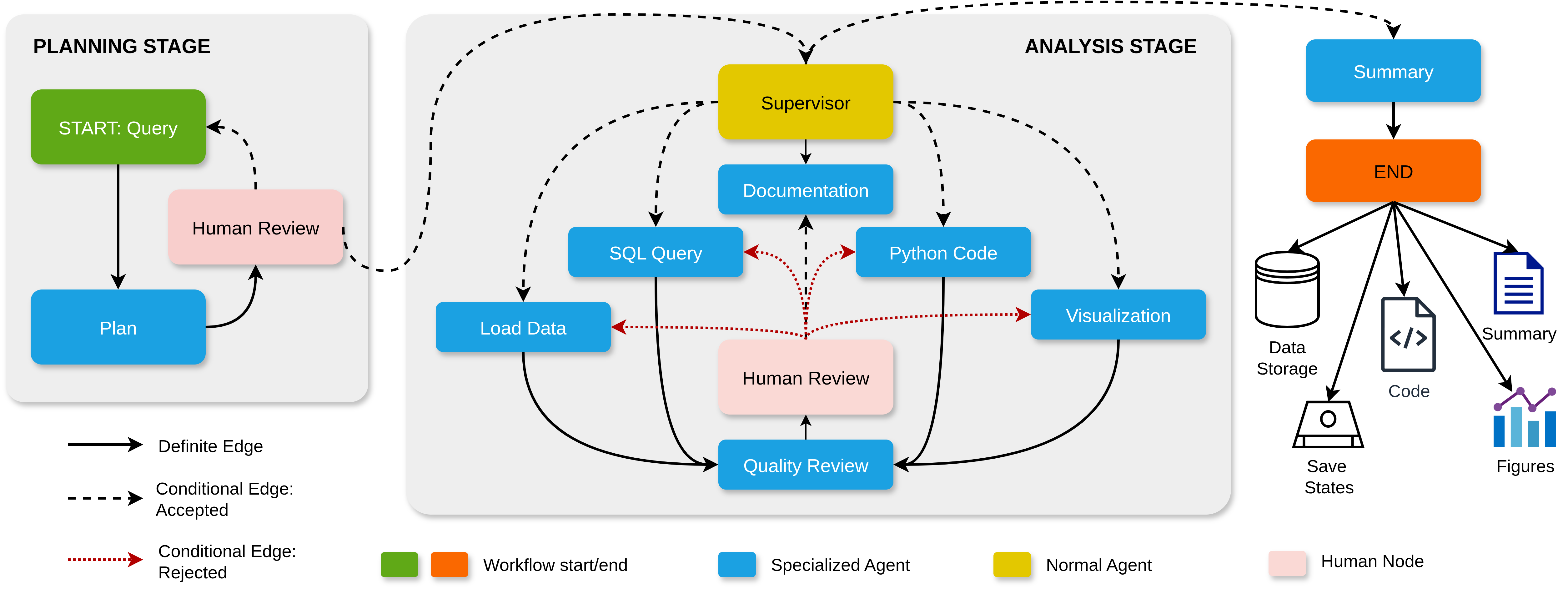
Automatic Explanation of Protein–Protein Binding Mechanism: A Preliminary Study
Computational Structural Bioinformatics Workshop, Springer Nature, 2025
This work explores how machine learning models can move beyond predicting binding affinity changes to automatically generating explanations of *why* a mutation affects a protein–protein interaction.
The study combines graph-based representations of protein interfaces with literature-derived annotations to associate model outputs with known biochemical mechanisms, serving as an early step toward interpretable mutation-effect prediction systems.
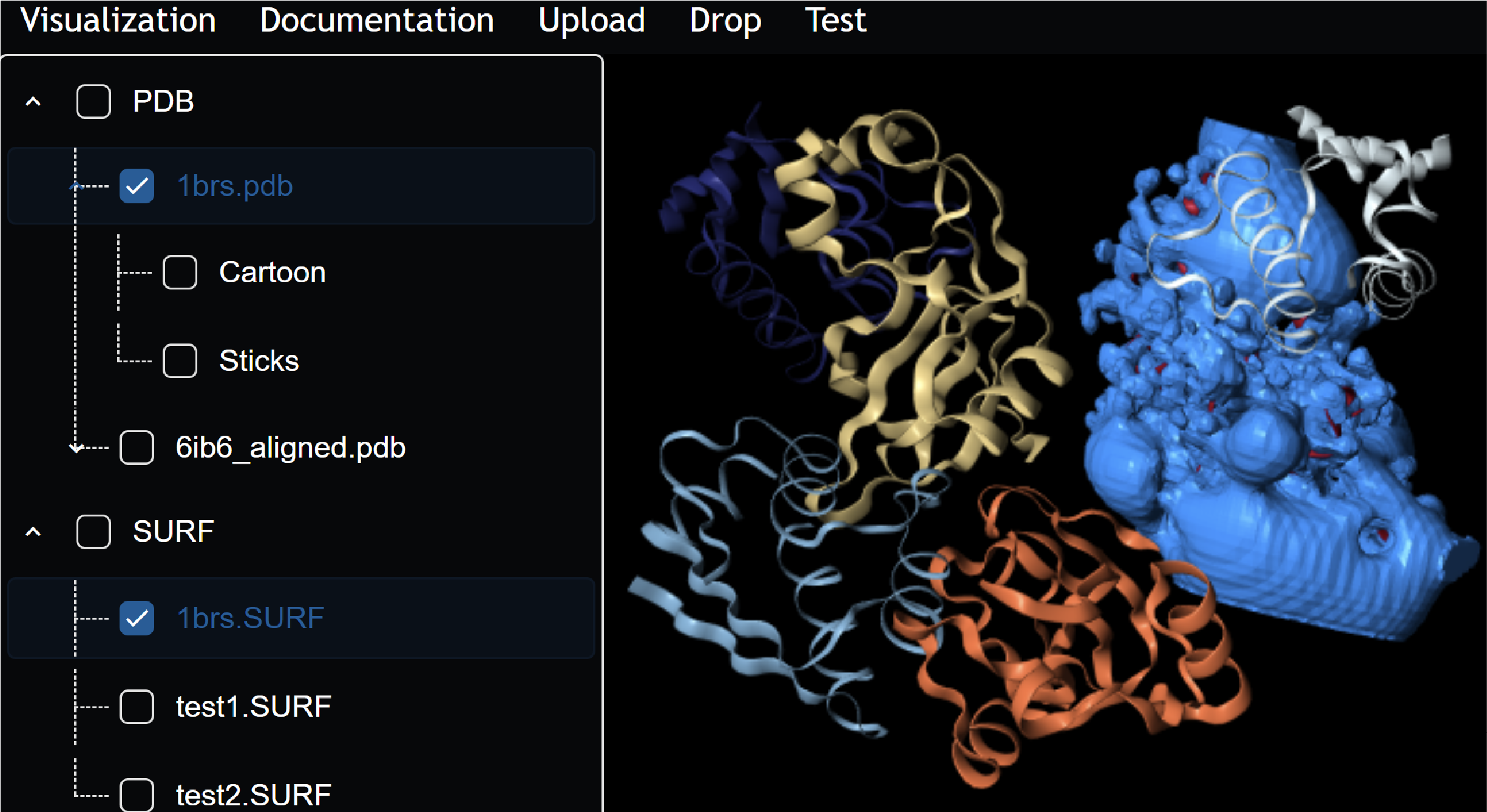
A Containerization Framework for Bioinformatics Software to Advance Scalability, Portability, and Maintainability
ACM International Conference on Bioinformatics, Computational Biology and Health Informatics (BCB ’23)
This paper introduces a framework for packaging and deploying bioinformatics tools as reusable, modular web services, lowering the barrier for researchers to build interactive and scalable scientific applications.
The work focuses on full-stack design, container orchestration, and abstraction layers that separate domain logic from infrastructure concerns. The framework was recognized as a Best Paper Finalist at IEEE CSBW and has been used to support multiple downstream research tools.
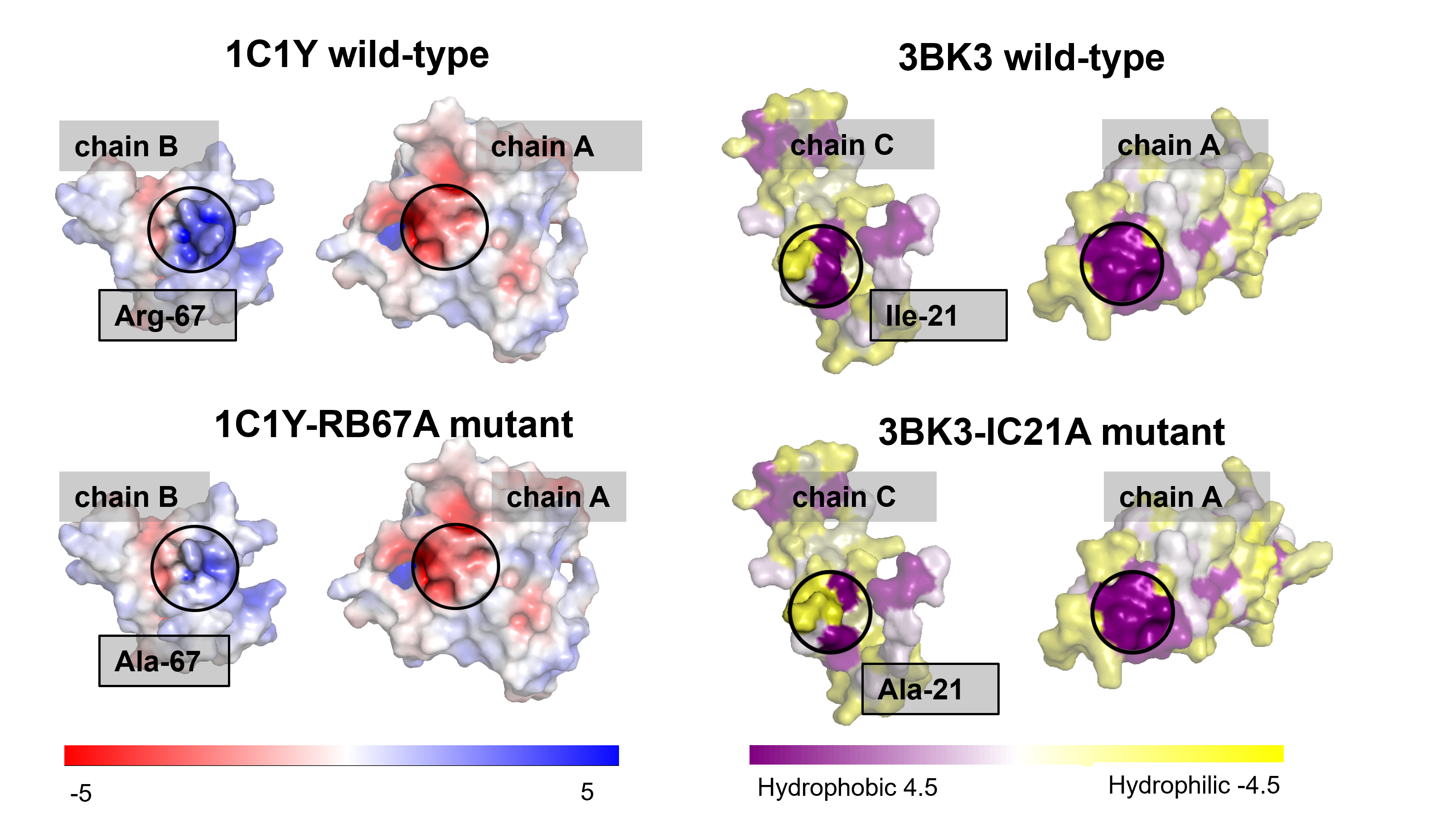
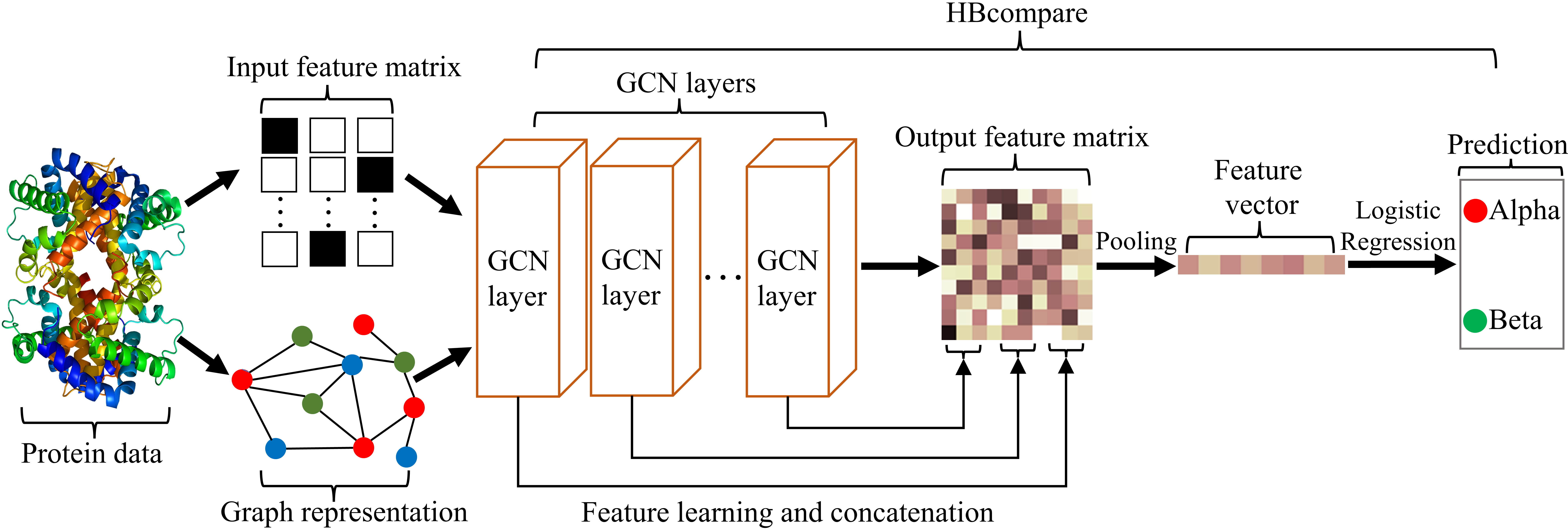
HBcompare: Classifying Ligand Binding Preferences with Hydrogen Bond Topology
Biomolecules, MDPI, 2022
HBcompare investigates how hydrogen bond topology influences ligand binding preferences across protein complexes.
The work introduces graph-based features that encode hydrogen bond patterns and uses these representations to classify binding behavior, demonstrating that topological structure alone can capture meaningful biochemical signals.
Analysis of Protein–Protein Interactions for Intermolecular Bond Prediction
Molecules, MDPI, 2022
This paper studies protein–protein interactions by predicting specific intermolecular bonds formed at binding interfaces using structural information.
The approach maps three-dimensional coordinate data to candidate bond interactions and validates predictions using experimentally derived mutation datasets, providing a foundation for later bond-network–based modeling.
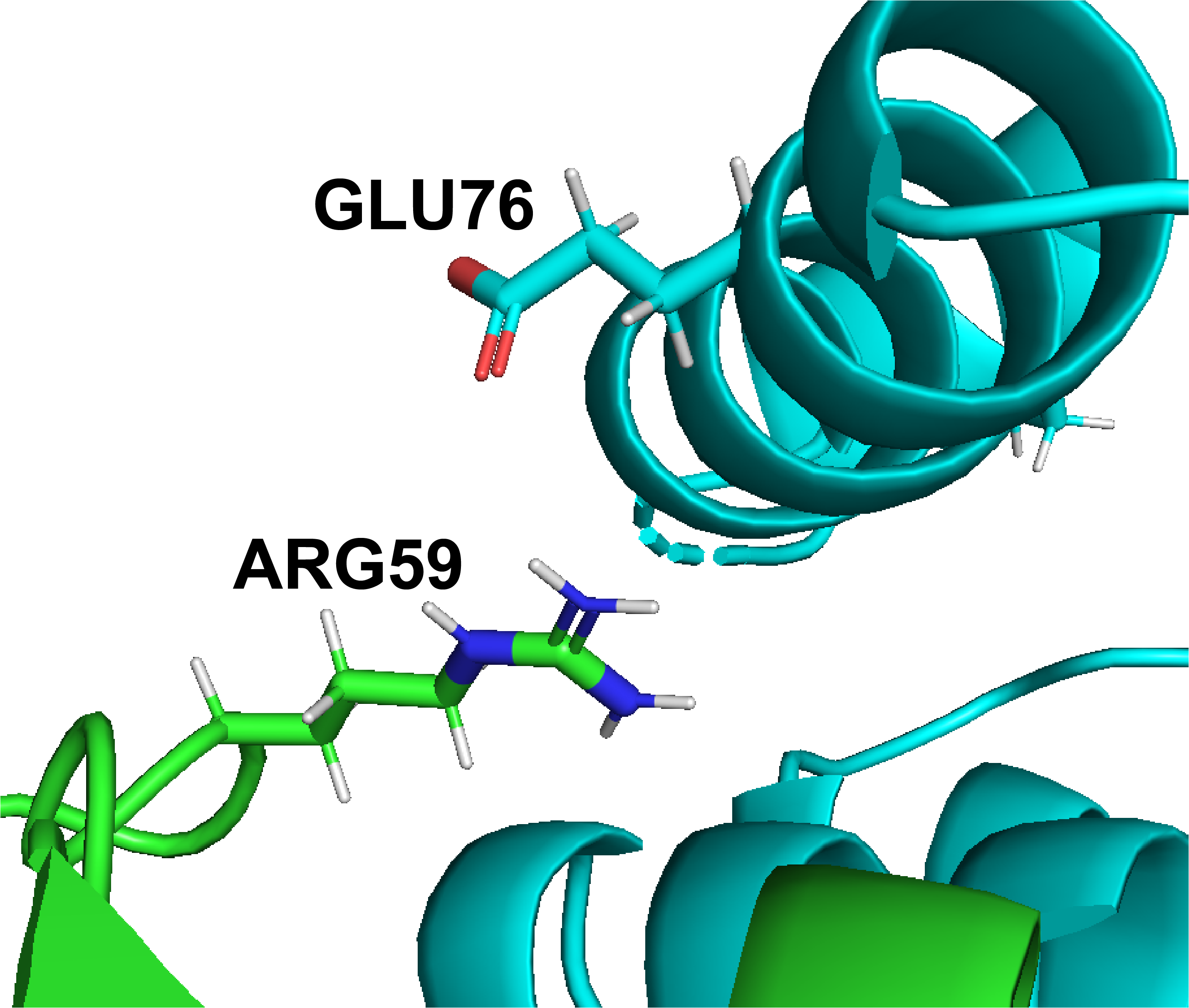
DiffBond: A Method for Predicting Intermolecular Bond Formation
IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2021
DiffBond introduces an early framework for predicting which intermolecular bonds form between interacting proteins based on structural features.
This work explores bond topology as a primary modeling target and laid the groundwork for later graph-based representations of protein interaction interfaces.

Education
PhD Candidate in Computer Science
Bioinformatics/Applied Machine Learning
2020 - 2026
Lehigh University, Bethlehem, PA
P.C. Rossin College of Engineering
Advisor: Brian Y. Chen
B.A. in Mathematics, Minor in Computer Science and Chemistry
2015 - 2019
Skidmore College, Saratoga Springs, NY


